Epitope Selection Software
Last December, the newly minted Parker Institute for Cancer Immunotherapy and its venerable East Coast counterpart, the Cancer Research Institute, announced the formation of the Tumor Neoantigen Selection Alliance. This initiative, involving researchers from 30 universities, non-profit institutions and companies, aims to identify software that can best predict mutation-associated cancer antigens, also known as neoantigens, from patient tumor DNA. The hope is that solving the shortcomings of current in silico methods for identifying neoantigens will galvanize a new wave of personalized cancer immunotherapies. But, for now, computational prediction of neoantigens capable of eliciting efficacious antitumor responses in patients remains a hit-or-miss affair. Cancer vaccines have traditionally targeted tumor-associated self-antigens—proteins that may be aberrantly expressed in cancer cells. More recently, however, attention has shifted to neoantigens. Targeting an individual's tumor-specific mutations is attractive because these peptides are new to the immune system and are not found in normal tissues.
The two figures vividly illustrate that selecting an epitope solely based on software algorithms is an essentially impossible task. Three different prediction programs, taken as an example, suggest a variety of different regions that might be suitable as an antigenic peptide. In contrast, the epitope mapping experiment. Lineage 1 Bot Programmiren there.
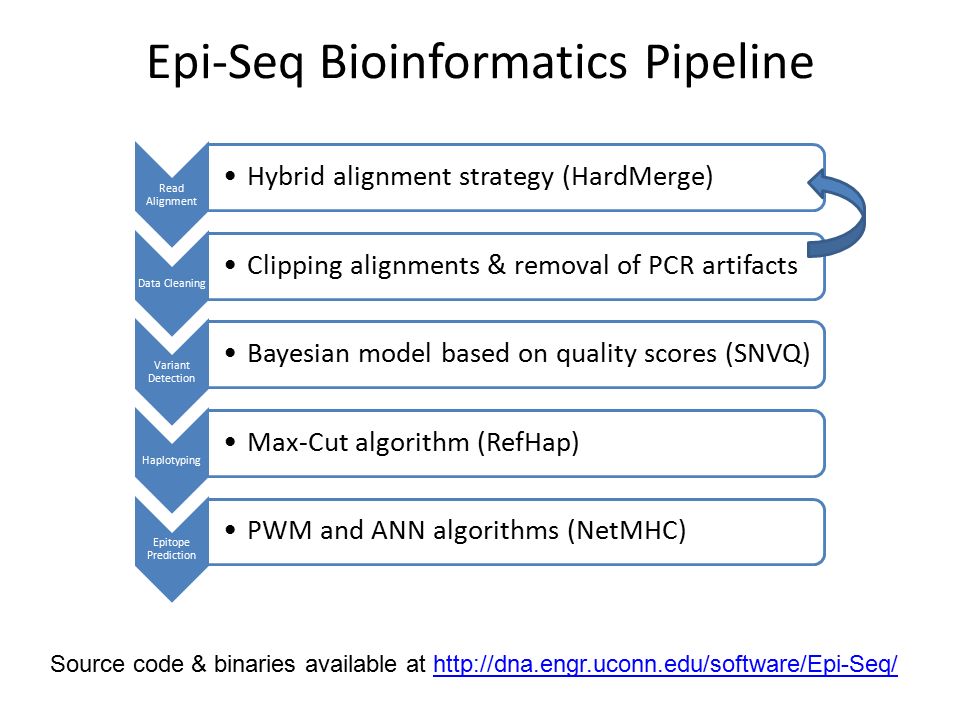
Compared with tumor-associated self-antigens, neoantigens elicit T-cell responses not subject to host central tolerance in the thymus and also produce fewer toxicities arising from autoimmune reactions to non-malignant cells. Although some computational methods focus on predicting what happens during antigen processing (e.g., NetChop) and peptide transport (e.g., NetCTL), most efforts focus on modeling which peptides bind to the MHC-I molecule. Neural network–based methods, such as NetMHC, are used to predict antigen sequences that generate epitopes fitting the groove of a patient's MHC-I molecules. Other filters can be applied to deprioritize hypothetical proteins and gauge whether a mutated amino acid either is likely orientated facing out of the MHC (toward the T-cell receptor) or reduces the affinity of the epitope for the MHC-I molecule itself. All kinds of confounding factors mean that these predictions can go awry.
Sequencing already introduces amplification biases and technical errors in the reads used as starting material for peptides. Modeling epitope processing and presentation also must take into account the fact that humans have ∼5,000 alleles encoding MHC-I molecules, with an individual patient expressing as many as six of them, all with different epitope affinities.
Methods such as NetMHC typically require 50–100 experimentally determined peptide-binding measurements for a particular allele to build a model with sufficient accuracy. But as many MHC alleles lack such data, 'pan-specific' methods—capable of predicting binders based on whether MHC alleles with similar contact environments have similar binding specificities—have increasingly come to the fore.
Background Reliable prediction of antibody, or B-cell, epitopes remains challenging yet highly desirable for the design of vaccines and immunodiagnostics. A correlation between antigenicity, solvent accessibility, and flexibility in proteins was demonstrated. Subsequently, Thornton and colleagues proposed a method for identifying continuous epitopes in the protein regions protruding from the protein's globular surface. The aim of this work was to implement that method as a web-tool and evaluate its performance on discontinuous epitopes known from the structures of antibody-protein complexes. Results Here we present ElliPro, a web-tool that implements Thornton's method and, together with a residue clustering algorithm, the MODELLER program and the Jmol viewer, allows the prediction and visualization of antibody epitopes in a given protein sequence or structure.
ElliPro has been tested on a benchmark dataset of discontinuous epitopes inferred from 3D structures of antibody-protein complexes. In comparison with six other structure-based methods that can be used for epitope prediction, ElliPro performed the best and gave an AUC value of 0.732, when the most significant prediction was considered for each protein.
Since the rank of the best prediction was at most in the top three for more than 70% of proteins and never exceeded five, ElliPro is considered a useful research tool for identifying antibody epitopes in protein antigens. Download When God Was A Woman Merlin Stone Pdf To Jpg there. ElliPro is available.
An antibody epitope, aka B-cell epitope or antigenic determinant, is a part of an antigen recognized by either a particular antibody molecule or a particular B-cell receptor of the immune system [ ]. For a protein antigen, an epitope may be either a short peptide from the protein sequence, called a continuous epitope, or a patch of atoms on the protein surface, called a discontinuous epitope. While continuous epitopes can be directly used for the design of vaccines and immunodiagnostics, the objective of discontinuous epitope prediction is to design a molecule that can mimic the structure and immunogenic properties of an epitope and replace it either in the process of antibody production–in this case an epitope mimic can be considered as a prophylactic or therapeutic vaccine–or antibody detection in medical diagnostics or experimental research [, ]. If continuous epitopes can be predicted using sequence-dependent methods built on available collections of immunogenic peptides (for review see [ ]), discontinuous epitopes–that are mostly the case when a whole protein, pathogenic virus, or bacteria is recognized by the immune system–are difficult to predict or identify from functional assays without knowledge of a three-dimensional (3D) structure of a protein [, ]. The first attempts at epitope prediction based on 3D protein structure began in 1984 when a correlation was established between crystallographic temperature factors and several known continuous epitopes of tobacco mosaic virus protein, myoglobin and lysozyme [ ]. A correlation between antigenicity, solvent accessibility, and flexibility of antigenic regions in proteins was also found [ ].